Abstract
Research Article
Pefloxacin and its derivative, novel inhibitors of the SARS-CoV-2 Main protease (3CLpro) and their pharmacokinetics prediction: An in silico analysis
Emmanuel Oluwaseun Adediran*
Published: 12 July, 2022 | Volume 6 - Issue 1 | Pages: 013-018
For over two years, COVID-19 pandemic has been a major global health concern and threat to human life. In the SARS-CoV2 macromolecules, the 3-chymotrypsin like protease (3CLpro or main protease) has been identified to be crucial and essential for viral survival, processing of the viral polyproteins and has been explored as a target in COVID-19 drug discovery.
Although vaccines and other various inhibitors have been designed and launched, the emergence of the variant of this virus has put an unrelenting effort of researchers to this end. Also, the high cost of manufacturing these molecules coupled with the occurrence of drug resistance is a concern.
Herein, Pefloxacin and its derivative for the first time were screened for their inhibitory activity against the SARS-CoV2 main protease through in silico analysis and their pharmacokinetic properties were evaluated. Interestingly, from the docking results, they both bind with high affinity at the active site of the protein. Moreover, they showed excellent pharmacokinetic and drug - likeness properties. Derivatization of Pefloxacin at the C7 position prevents its blood-brain barrier permeability.
Overall, the dual antibacterial and potential antiviral activities of these two molecules make them promising drug candidates for COVID-19 management.
Read Full Article HTML DOI: 10.29328/journal.apps.1001030 Cite this Article Read Full Article PDF
Keywords:
Pefloxacin; 1-ethyl-6-fluro-7-(4(Nsubstitutedcarbamoylmethylphenyl)piperazyn-1-yl)-4-oxoquinoline-3-carboxylic acid; SARS-CoV2; Main-protease(3CLpro); Docking
References
- Correia S, Poeta P, Hébraud M, Capelo JL, Igrejas G. Mechanisms of quinolone action and resistance: where do we stand? J Med Microbiol. 2017 May;66(5):551-559. doi: 10.1099/jmm.0.000475. Epub 2017 May 12. PMID: 28504927.
- Suaifan GARY, Mohammed AAM. Fluoroquinolones structural and medicinal developments (2013-2018): Where are we now? Bioorg Med Chem. 2019 Jul 15;27(14):3005-3060. doi: 10.1016/j.bmc.2019.05.038. Epub 2019 May 31. Erratum in: Bioorg Med Chem. 2019 Nov 1;27(21):115072. PMID: 31182257.
- Richter S, Parolin C, Palumbo M, Palù G. Antiviral properties of quinolone-based drugs. Curr Drug Targets Infect Disord. 2004 Jun;4(2):111-6. doi: 10.2174/1568005043340920. PMID: 15180459.
- Ali SH, Chandraker A, DeCaprio JA. Inhibition of Simian virus 40 large T antigen helicase activity by fluoroquinolones. Antivir Ther. 2007;12(1):1-6. PMID: 17503741.
- Fedorowicz J, Sączewski J. Modifications of quinolones and fluoroquinolones: hybrid compounds and dual-action molecules. Monatsh Chem. 2018;149(7):1199-1245. doi: 10.1007/s00706-018-2215-x. Epub 2018 Jun 7. PMID: 29983452; PMCID: PMC6006264.
- Richter S, Parolin C, Palumbo M, Palù G. Antiviral properties of quinolone-based drugs. Curr Drug Targets Infect Disord. 2004 Jun;4(2):111-6. doi: 10.2174/1568005043340920. PMID: 15180459.
- Marciniec K, Beberok A, Pęcak P, Boryczka S, Wrześniok D. Ciprofloxacin and moxifloxacin could interact with SARS-CoV-2 protease: preliminary in silico Pharmacol Rep. 2020 Dec;72(6):1553-1561. doi: 10.1007/s43440-020-00169-0. Epub 2020 Oct 15. PMID: 33063271; PMCID: PMC7561236.
- Alexpandi R, De Mesquita JF, Pandian SK, Ravi AV. Quinolines-Based SARS-CoV-2 3CLpro and RdRp Inhibitors and Spike-RBD-ACE2 Inhibitor for Drug-Repurposing Against COVID-19: An in silico Front Microbiol. 2020 Jul 23;11:1796. doi: 10.3389/fmicb.2020.01796. PMID: 32793181; PMCID: PMC7390959.
- GhebreyesusTA “WHO Director-General’s opening remarks at the media briefing on COVID-19 World Heal Organ”. 2020; 4.
- Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020 Apr 24;368(6489):409-412. doi: 10.1126/science.abb3405. Epub 2020 Mar 20. PMID: 32198291; PMCID: PMC7164518.
- Khan RJ, Jha RK, Amera GM, Jain M, Singh E, Pathak A, Singh RP, Muthukumaran J, Singh AK. Targeting SARS-CoV-2: a systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2'-O-ribose methyltransferase. J Biomol Struct Dyn. 2021 May;39(8):2679-2692. doi: 10.1080/07391102.2020.1753577. Epub 2020 Apr 20. PMID: 32266873; PMCID: PMC7189412.
- Fearon D, Powell AJ, Douangamath A, Owen CD, Wild C, Krojer T, et al. PanDDA analysis group deposition—crystal structure of COVID-19 main protease in complex with Z1220452176.
- Liu X, Zhang B, Jin Z, Yang H, Rao Z. The crystal structure of COVID-19 main protease in complex with an inhibitor N3. 2020. https://doi.org/10.2210/pdb6LU7/pdb.
- Santana MVS, Silva-Jr FP. De novo design and bioactivity prediction of SARS-CoV-2 main protease inhibitors using recurrent neural network-based transfer learning. BMC Chem. 2021 Feb 2;15(1):8. doi: 10.1186/s13065-021-00737-2. PMID: 33531083; PMCID: PMC7852053.
- Saakre M, Mathew D, Ravisankar V. Perspectives on plant flavonoid quercetin-based drugs for novel SARS-CoV-2. Beni Suef Univ J Basic Appl Sci. 2021;10(1):21. doi: 10.1186/s43088-021-00107-w. Epub 2021 Mar 24. PMID: 33782651; PMCID: PMC7989718.
- Das S, Sarmah S, Lyndem S, Singha Roy A. An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. J Biomol Struct Dyn. 2021 Jun;39(9):3347-3357. doi: 10.1080/07391102.2020.1763201. Epub 2020 May 13. PMID: 32362245; PMCID: PMC7232884.
- M Khan MT, Ali A, Wang Q, Irfan M, Khan A, Zeb MT, Zhang YJ, Chinnasamy S, Wei DQ. Marine natural compounds as potents inhibitors against the main protease of SARS-CoV-2-a molecular dynamic study. J Biomol Struct Dyn. 2021 Jul;39(10):3627-3637. doi: 10.1080/07391102.2020.1769733. Epub 2020 Jun 1. PMID: 32410504; PMCID: PMC7284144.
- Joshi T, Sharma P, Joshi T, Pundir H, Mathpal S, Chandra S. Structure-based screening of novel lichen compounds against SARS Coronavirus main protease (Mpro) as potentials inhibitors of COVID-19. Mol Divers. 2021 Aug;25(3):1665-1677. doi: 10.1007/s11030-020-10118-x. Epub 2020 Jun 29. PMID: 32602074; PMCID: PMC7323881.
- Sk MF, Roy R, Jonniya NA, Poddar S, Kar P. Elucidating biophysical basis of binding of inhibitors to SARS-CoV-2 main protease by using molecular dynamics simulations and free energy calculations. J Biomol Struct Dyn. 2021 Jul;39(10):3649-3661. doi: 10.1080/07391102.2020.1768149. Epub 2020 Jun 1. PMID: 32396767; PMCID: PMC7284146.
- Bhardwaj VK, Singh R, Sharma J, Rajendran V, Purohit R, Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J Biomol Struct Dyn. 2021 Jul;39(10):3449-3458. doi: 10.1080/07391102.2020.1766572. Epub 2020 May 20. PMID: 32397940; PMCID: PMC7256349.
- Qu H, Zheng Y, Wang Y, Li H, Liu X, Xiong X, Zhang L, Gu J, Yang G, Zhu Z, Zheng H, Ouyang Q. The potential effects of clinical antidiabetic agents on SARS-CoV-2. J Diabetes. 2021 Mar;13(3):243-252. doi: 10.1111/1753-0407.13135. Epub 2020 Dec 19. PMID: 33210826; PMCID: PMC7753367.
- Rabie AM. RETRACTED ARTICLE: Discovery of (E)-N-(4-cyanobenzylidene)-6-fluoro-3-hydroxypyrazine-2-carboxamide (cyanorona-20): the first potent and specific anti-COVID-19 drug. Chem Zvesti. 2021;75(9):4669-4685. doi: 10.1007/s11696-021-01640-9. Epub 2021 May 16. Retraction in: Chem Zvesti. 2022 Jun 4;:1. PMID: 34025012; PMCID: PMC8126404.
- Gao H, Sun X, Rao Y. PROTAC Technology: Opportunities and Challenges. ACS Med Chem Lett. 2020 Mar 12;11(3):237-240. doi: 10.1021/acsmedchemlett.9b00597. PMID: 32184950; PMCID: PMC7073876.
- Di Sarno V, Lauro G, Musella S, Ciaglia T, Vestuto V, Sala M, Scala MC, Smaldone G, Di Matteo F, Novi S, Tecce MF, Moltedo O, Bifulco G, Campiglia P, Gomez-Monterrey IM, Snoeck R, Andrei G, Ostacolo C, Bertamino A. Identification of a dual acting SARS-CoV-2 proteases inhibitor through in silico design and step-by-step biological characterization. Eur J Med Chem. 2021 Dec 15;226:113863. doi: 10.1016/j.ejmech.2021.113863. Epub 2021 Sep 22. PMID: 34571172; PMCID: PMC8457654.
- Andrzejczyk, J, Jovic, K, Brown, LM, Pascetta, VG, Varga, K, Vashisth, H(2022). Molecular interactions and inhibition of the SARS-CoV-2 main protease by a thiazolidinedione derivative. Proteins; 1- 12.https:// doi/1002/prot.26385
- Mediouni S, Mou H, Otsuka Y, Jablonski JA, Adcock RS, Batra L, Chung DH, Rood C, de Vera IMS, Rahaim R Jr, Ullah S, Yu X, Getmanenko YA, Kennedy NM, Wang C, Nguyen TT, Hull M, Chen E, Bannister TD, Baillargeon P, Scampavia L, Farzan M, Valente ST, Spicer TP. Identification of potent small molecule inhibitors of SARS-CoV-2 entry. SLAS Discov. 2022 Jan;27(1):8-19. doi: 10.1016/j.slasd.2021.10.012. Epub 2021 Oct 23. PMID: 35058179; PMCID: PMC8577999.
- Singh P, Chauhan SS, Pandit S, Sinha M, Gupta S, Gupta A, Parthasarathi R. The dual role of phytochemicals on SARS-CoV-2 inhibition by targeting host and viral proteins. J Tradit Complement Med. 2022 Jan;12(1):90-99. doi: 10.1016/j.jtcme.2021.09.001. Epub 2021 Sep 8. PMID: 34513611; PMCID: PMC8424525.
- Tang B, He F, Liu D, He F, Wu T, Fang M, Niu Z, Wu Z, Xu D. AI-Aided Design of Novel Targeted Covalent Inhibitors against SARS-CoV-2. Biomolecules. 2022; 12(6):746. https://doi.org/10.3390/biom12060746
- Benítez-Cardoza CG, Vique-Sánchez JL. Potential inhibitors of the interaction between ACE2 and SARS-CoV-2 (RBD), to develop a drug. Life Sci. 2020 Sep 1;256:117970. doi: 10.1016/j.lfs.2020.117970. Epub 2020 Jun 15. PMID: 32553928; PMCID: PMC7294299.
- Khaerunnisa S, Kurniawan H, Awaluddin R, Suhartati S, Soetjipto S. Potential Inhibitor of COVID- 19 Main Protease (Mpro) from Several Medicinal Plant Compounds by Molecular Docking Study. Preprints. 2020. doi: 10.20944/preprints202003.0226.v1
- Dallakyan S, Olson AJ. Small-Molecule Library Screening by Docking with PyRx. In: Hempel, J., Williams, ., Hong, C. (eds) Chemical Biology. Methods in Molecular Biology. 2015; 1263. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-2269-7_19
- Roymans D, De Bondt HL, Arnoult E, Geluykens P, Gevers T, Van Ginderen M, Verheyen N. Binding of a potent small-molecule inhibitor of six-helix bundle formation require interactions with both heptad-repeats of the RSV fusion protein. PNAS https://doi.org/10.1073/pnas.091010810
- Al-Zrkani MK, Abdulkareem RA, Al-Fahad D, Al Shouber M, Nasr AMS, Al-Khdhairawi A. Elucidating novel antibacterial compounds from the NPASS database against the FimH lectin domain for the treatment of urinary tract infections: an in-silico study. J Biomol Struct Dyn. 2022 Apr 9:1-12. doi: 10.1080/07391102.2022.2059009. Epub ahead of print. PMID: 35403563.
- Rauf A, Raza M, Humayun Khan M, Hemeg HA, Al-Awthan YS, Bahattab O, Bawazeer S, Naz S, Basoglu F, Saleem M, Khan M, Seyyedamirhossein H, Mubarak MS, Erdogan Orhan I. In vitroand in silico studies on clinically important enzymes inhibitory activities of flavonoids isolated from Euphorbia pulcherrima. Ann Med. 2022 Jan 27;54(1):495-506. doi: 10.1080/07853890.2022.2033826. PMID: 35112936; PMCID: PMC8820783.
- Goodsell DS, Morris GM, Olson AJ. Automated docking of flexible ligands: applications of AutoDock. J Mol Recognit. 1996 Jan-Feb;9(1):1-5. doi: 10.1002/(sici)1099-1352(199601)9:1<1::aid-jmr241>3.0.co;2-6. PMID: 8723313.
- Lipinski CA. Drug-like properties and the causes of poor solubility and poor permeability. J Pharmacol Toxicol Methods. 2000 Jul-Aug;44(1):235-49. doi: 10.1016/s1056-8719(00)00107-6. PMID: 11274893.
- Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017 Mar 3;7:42717. doi: 10.1038/srep42717. PMID: 28256516; PMCID: PMC5335600.
- Mustafa M, Mostafa YA. Antimicrobial Pyridazines: Synthesis, Characterization, Cytotoxicity, Substrate Promiscuity, and Molecular Docking. Chem Biodivers. 2020 Jun;17(6):e2000100. doi: 10.1002/cbdv.202000100. Epub 2020 May 8. PMID: 32239712.
- Khadse SC, Amnerkar ND, Dave MU.Quinazolin-4-one derivatives lacking toxicity-producing attributes as glucokinase activators: design, synthesis, molecular docking, and in-silico ADMET prediction. Fut J Pharm Sci; 2019; 5: 11.https://doi.org/10.1186/s43094-019-0012-y
Figures:
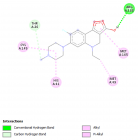
Figure 1
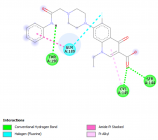
Figure 2
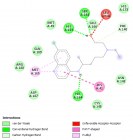
Figure 3
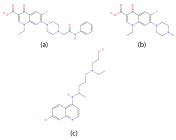
Figure 4
Similar Articles
-
Pefloxacin and its derivative, novel inhibitors of the SARS-CoV-2 Main protease (3CLpro) and their pharmacokinetics prediction: An in silico analysisEmmanuel Oluwaseun Adediran*. Pefloxacin and its derivative, novel inhibitors of the SARS-CoV-2 Main protease (3CLpro) and their pharmacokinetics prediction: An in silico analysis. . 2022 doi: 10.29328/journal.apps.1001030; 6: 013-018
Recently Viewed
-
Eye Banking a Boon in Eye Health Care Services and Vision Restoration and Corneal Tattooing for Aesthetic PurposesShripat Dixit*, Reva Kanash, Hema Joshi. Eye Banking a Boon in Eye Health Care Services and Vision Restoration and Corneal Tattooing for Aesthetic Purposes. Int J Clin Exp Ophthalmol. 2023: doi: 10.29328/journal.ijceo.1001050; 7: 001-002
-
Exophthalmos Revealing a Spheno Temporo Orbital MeningiomaHassina S*, Krichene MA, Hazil Z, Bekkar B, Hasnaoui I, Robbana L, Bardi S, Akkanour Y, Serghini L, Abdallah EL. Exophthalmos Revealing a Spheno Temporo Orbital Meningioma. Int J Clin Exp Ophthalmol. 2024: doi: 10.29328/journal.ijceo.1001055; 8: 001-003
-
Evaluation of the health of temporomandibular joint – A novel techniqueSuguna Priya G*,Shwetha Poovani,Prafulla Thumati. Evaluation of the health of temporomandibular joint – A novel technique. J Oral Health Craniofac Sci. 2022: doi: 10.29328/journal.johcs.1001037; 7: 005-009
-
Periodontal suturing, data on selection criteriaIlma Robo*,Saimir Heta,Migerta Cafaj,Sonila Robo,Eduart Kapaj. Periodontal suturing, data on selection criteria. J Oral Health Craniofac Sci. 2022: doi: 10.29328/journal.johcs.1001038; 7: 010-013
-
Current and emerging trends in oro-dental healthcare and cranio-maxillo-facial surgeryZiyad S Haidar*. Current and emerging trends in oro-dental healthcare and cranio-maxillo-facial surgery. J Oral Health Craniofac Sci. 2023: doi: 10.29328/journal.johcs.1001042; 8: 001-006
Most Viewed
-
Feasibility study of magnetic sensing for detecting single-neuron action potentialsDenis Tonini,Kai Wu,Renata Saha,Jian-Ping Wang*. Feasibility study of magnetic sensing for detecting single-neuron action potentials. Ann Biomed Sci Eng. 2022 doi: 10.29328/journal.abse.1001018; 6: 019-029
-
Evaluation of In vitro and Ex vivo Models for Studying the Effectiveness of Vaginal Drug Systems in Controlling Microbe Infections: A Systematic ReviewMohammad Hossein Karami*, Majid Abdouss*, Mandana Karami. Evaluation of In vitro and Ex vivo Models for Studying the Effectiveness of Vaginal Drug Systems in Controlling Microbe Infections: A Systematic Review. Clin J Obstet Gynecol. 2023 doi: 10.29328/journal.cjog.1001151; 6: 201-215
-
Prospective Coronavirus Liver Effects: Available KnowledgeAvishek Mandal*. Prospective Coronavirus Liver Effects: Available Knowledge. Ann Clin Gastroenterol Hepatol. 2023 doi: 10.29328/journal.acgh.1001039; 7: 001-010
-
Causal Link between Human Blood Metabolites and Asthma: An Investigation Using Mendelian RandomizationYong-Qing Zhu, Xiao-Yan Meng, Jing-Hua Yang*. Causal Link between Human Blood Metabolites and Asthma: An Investigation Using Mendelian Randomization. Arch Asthma Allergy Immunol. 2023 doi: 10.29328/journal.aaai.1001032; 7: 012-022
-
An algorithm to safely manage oral food challenge in an office-based setting for children with multiple food allergiesNathalie Cottel,Aïcha Dieme,Véronique Orcel,Yannick Chantran,Mélisande Bourgoin-Heck,Jocelyne Just. An algorithm to safely manage oral food challenge in an office-based setting for children with multiple food allergies. Arch Asthma Allergy Immunol. 2021 doi: 10.29328/journal.aaai.1001027; 5: 030-037

HSPI: We're glad you're here. Please click "create a new Query" if you are a new visitor to our website and need further information from us.
If you are already a member of our network and need to keep track of any developments regarding a question you have already submitted, click "take me to my Query."